Resampling in SimpleITK
Oblique slices
 SimpleITK
SimpleITKSimpleITK has a fantastic python API for medical image processing, but it has a steep learning curve. In this post I will document how to resample images using the Resample filter via the complicated, but really useful task of extracting oblique slices in a 3D image. If you imagine a 3D image as a cube then an oblique slice is formed by sampling the image on a plane that has an arbitrary normal and does not align with the canonical axial, sagittal, coronal planes (see Fig. 1).
Figure 1 shows the typical setup in SimpleITK where the image field of view
is denoted by the dotted cube. Let
I am assuming that we are given
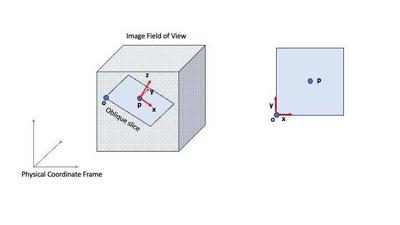
Given a SimpleITK img, I typically do the following to resample it:
resample = sitk.ResampleImageFilter()
resample.SetOutputDirection(output_direction)
resample.SetOutputOrigin(output_origin)
resample.SetOutputSpacing(output_spacing)
resample.SetSize(output_size)
resample.SetTransform(sitk.Transform())
resample.SetDefaultPixelValue(0)
resample.SetInterpolator(sitk.sitkLinear)
resample.Execute(img)
We have to specify the direction, origin, spacing, and size (in voxels) for the final resampled slice.
direction in a SimpleITK header is a (9, 1)-shaped array that expresses the rotation matrix (in a row-major form) that takes the physical reference frame (
Let the rotation matrix be
R = np.zeros((3,3))
R[:,0] = x
R[:,1] = y
R[:,2] = z
The SimpleITK direction part of the header of the output slice will therefore be,
direction =
output_direction = R.ravel()
or
output_direction_ = [x[0], y[0], z[0], x[1], y[1], z[1], x[2], y[2], z[2]]
To identify the physical coordinates of the origin
where the unit vector
u = (x + y)/np.sqrt(2)
d = (s * voxel_spacing)/np.sqrt(2)
output_origin = p - d*u
That’s about it. We can specify the rest of the output image parameters and
execute the resample filter to extract an oblique slice centered at a point
SimpleITK has a nice set of notebooks here and perhaps if I had done the homework exercises my learning curve would not have been so steep.